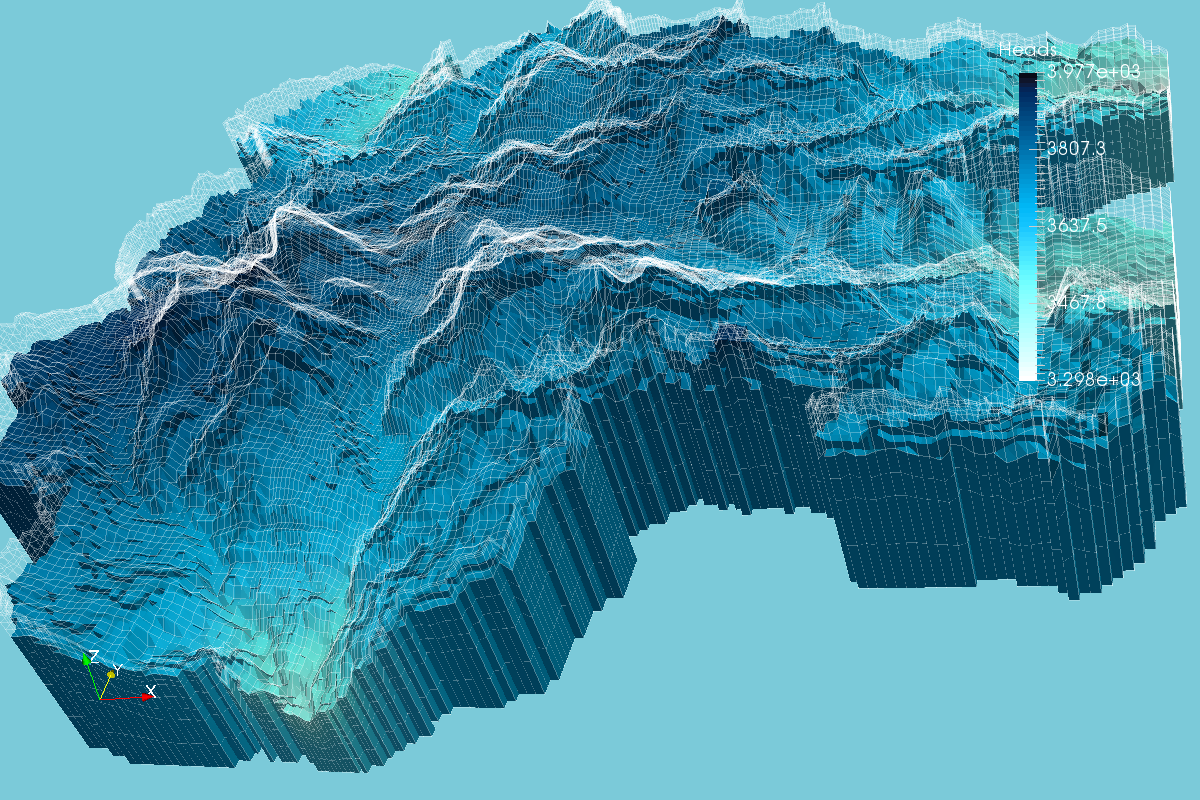
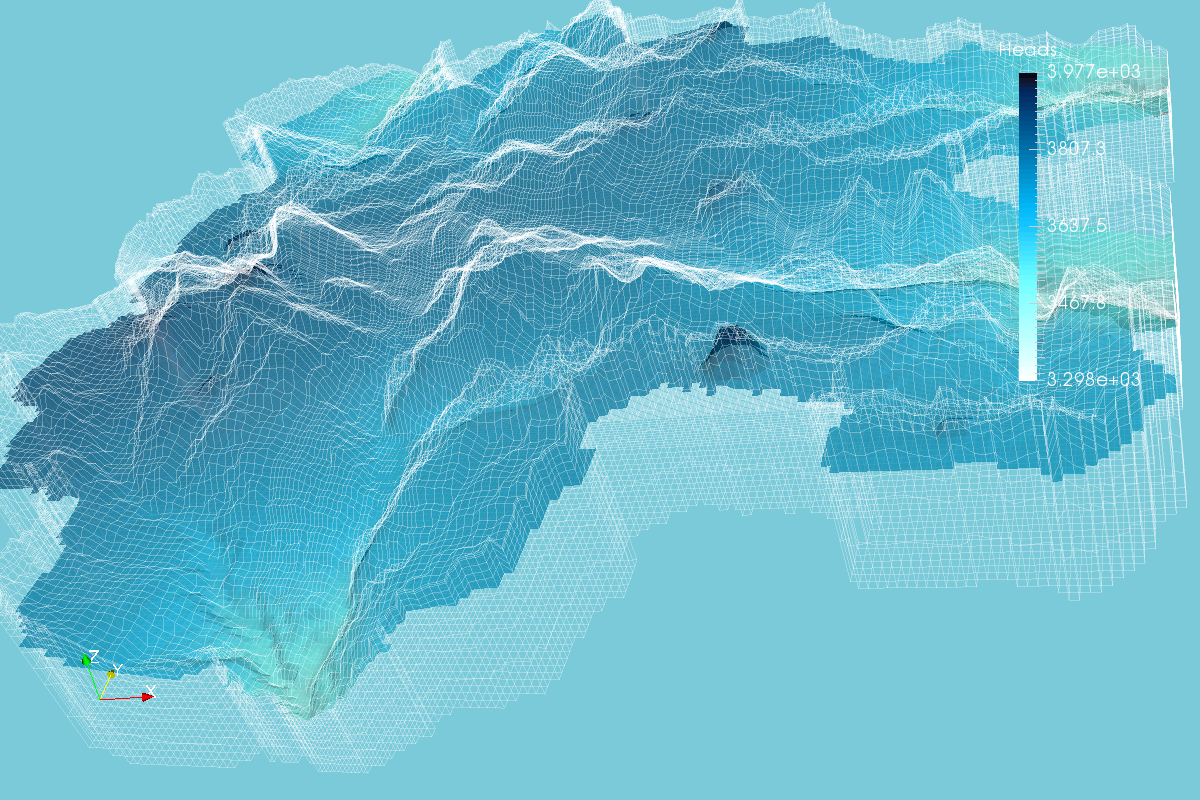
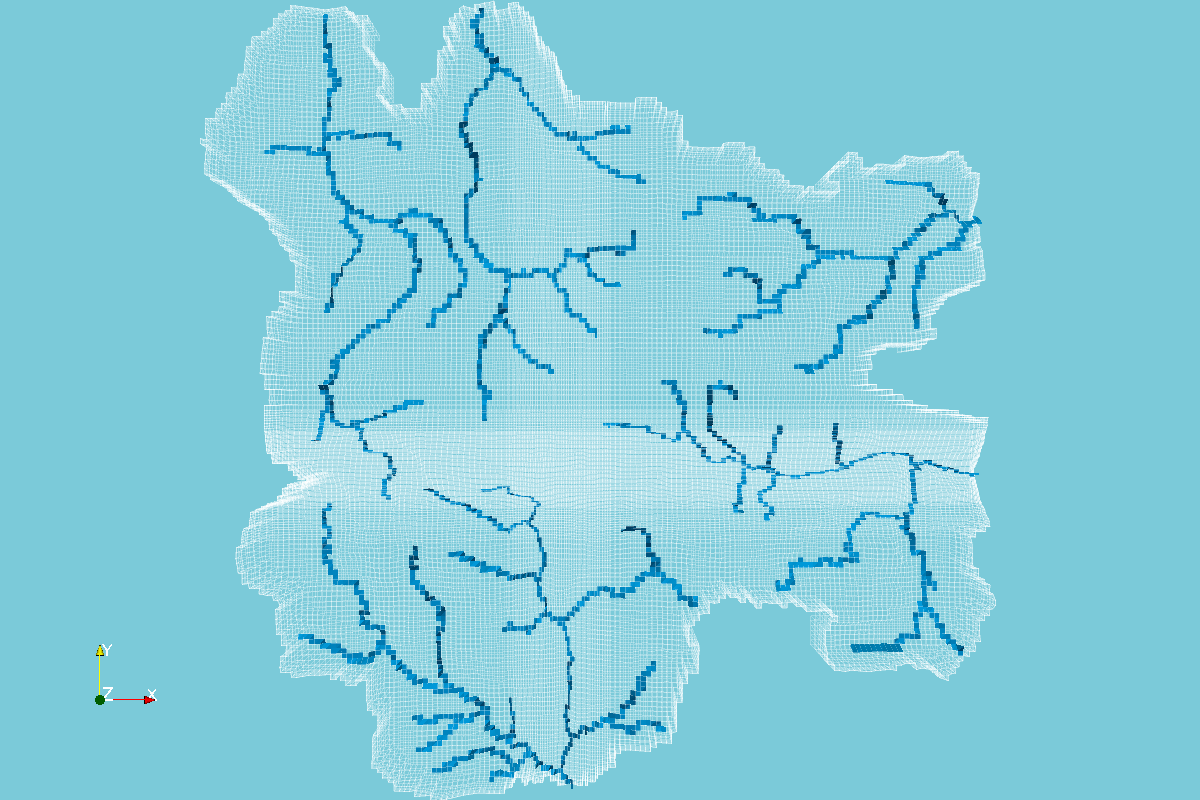
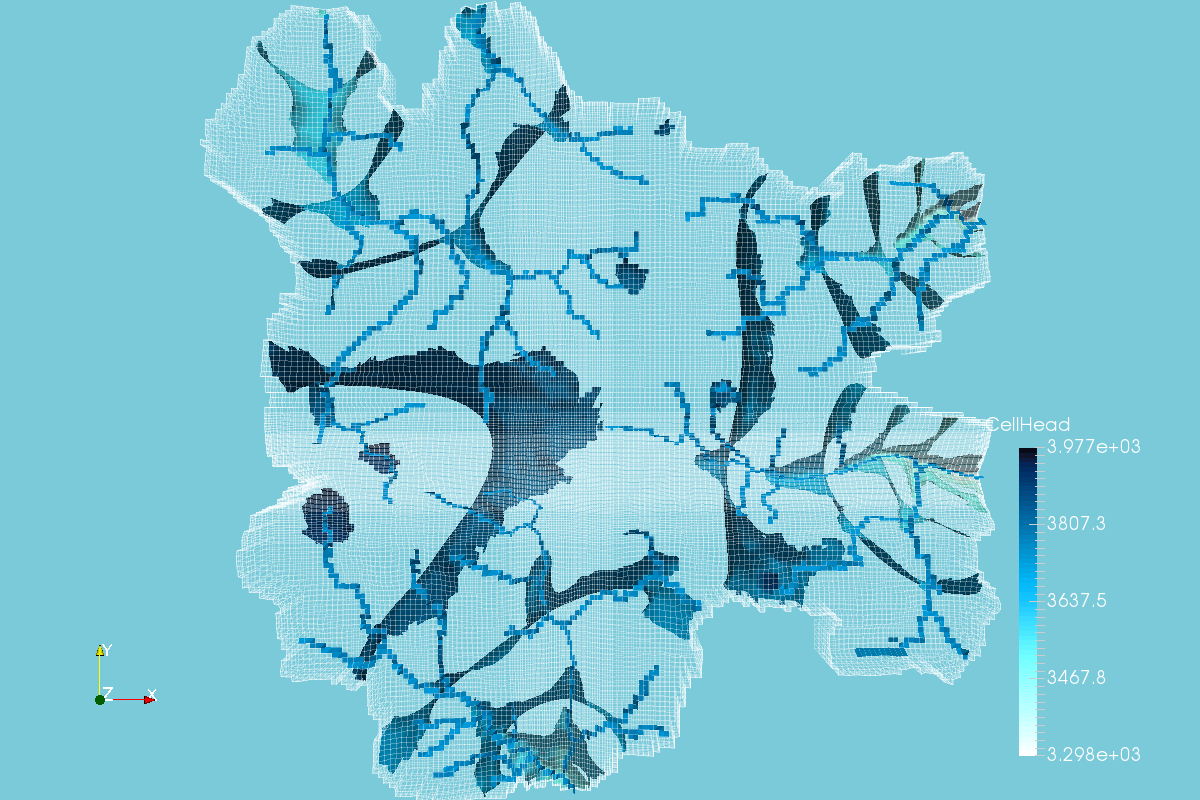
Enhanced MODFLOW Result Representation with Python, VTK and Paraview - Tutorial
/MODFLOW model output representation is key to understand the groundwater flow regime, to study the interaction with surface water and depending ecosystems and to evaluate the impact to anthropogenic and climate change requirements. Until now, there has been few open source software capable of generating 3D representations and those software had limited options for color scales, cross sections and other graphical tools. On the research for more options we found Paraview, a open source software for data representation designed to analyze extremely large datasets using distributed memory computing resources.
In order to represent MODFLOW output into Paraview, a VTK file for unstructured grids is needed, this VTK type is called VTU where the "U" comes from unestructured. The tutorial shows the complete procedure to process a MODFLOW model output into a VTU file and the representation in Paraview.
Photo Gallery
Tutorial
Input files
Files for this tutorial can be downloaded here
Code
This is the complete code used in this tutorial.
# coding: utf-8
# # Import the required packages and open model files
# In[1]:
# import the required libraries
import os, re
import numpy as np
from workingFunctions import Functions #functions from the workingFunctions.py file
# In[2]:
#change directory to the script path
os.chdir('C:/Users\Saul\Documents\Ih_PlayaroundwithVTK') #use your own path
#open the DIS, BAS and FHD and DRN files
disLines= open('Model/Modelo3.dis').readlines() #discretization data
basLines= open('Model/Modelo3.bas').readlines() #active / inactive data
fhdLines= open('Model/Modelo3.fhd').readlines() #water head data
drnLines= open('Model/Modelo3.drn').readlines() #drain package data
#create a empty dictionay to store the model features
modDis = {}
modBas = {}
modFhd = {}
modDrn = {}
# # Working with the DIS (Discretization Data) data
# ### General model features as modDis dict
# In[3]:
#get the extreme coordinates form the dis header
for line in disLines[:5]:
line = re.sub('\(|\)|,',' ',line) #change the (, ) and , character in the dis file
if "Lower left corner" in line:
line = line.split()
modDis["vertexXmin"]=float(line[4])
modDis["vertexYmin"]=float(line[5])
elif "Upper right corner" in line:
line = line.split()
modDis["vertexXmax"]=float(line[4])
modDis["vertexYmax"]=float(line[5])
#get the number of layers, rows, columns, cell and vertex numbers
linelaycolrow = disLines[6].split()
modDis["cellLays"] = int(linelaycolrow[0])
modDis["cellRows"] = int(linelaycolrow[1])
modDis["cellCols"] = int(linelaycolrow[2])
modDis["vertexLays"] = modDis["cellLays"] + 1
modDis["vertexRows"] = modDis["cellRows"] + 1
modDis["vertexCols"] = modDis["cellCols"] + 1
modDis["vertexperlay"] = modDis["vertexRows"] * modDis["vertexCols"]
modDis["cellsperlay"] = modDis["cellRows"] * modDis["cellCols"]
# ### Get the DIS Breakers
# In[4]:
#get the grid breakers
modDis['disBreakers']={}
breakerValues = ["INTERNAL","CONSTANT"]
vertexLay=0
for item in breakerValues:
for line in disLines:
if item in line:
if 'DELR' in line: #DELR is cell width along rows
modDis['disBreakers']['DELR']=disLines.index(line)
elif 'DELC' in line: #DELC is cell width along columns
modDis['disBreakers']['DELC']=disLines.index(line)
else:
modDis['disBreakers']['vertexLay'+str(vertexLay)]=disLines.index(line)
vertexLay+=1
# ### Get the DEL Info
# In[5]:
modDis['DELR'] = Functions.getListFromDEL(modDis['disBreakers']['DELR'],disLines,modDis['cellCols'])
modDis['DELC'] = Functions.getListFromDEL(modDis['disBreakers']['DELC'],disLines,modDis['cellRows'])
# ### Get the Cell Centroid Z
# In[6]:
modDis['cellCentroidZList']={}
for lay in range(modDis['vertexLays']):
#add auxiliar variables to identify breakers
lineaBreaker = modDis['disBreakers']['vertexLay'+str(lay)]
#two cases in breaker line
if 'INTERNAL' in disLines[lineaBreaker]:
lista = Functions.getListFromBreaker(lineaBreaker,modDis,disLines)
modDis['cellCentroidZList']['lay'+str(lay)]=lista
elif 'CONSTANT' in disLines[lineaBreaker]:
constElevation = float(disLines[lineaBreaker].split()[1])
modDis['cellCentroidZList']['lay'+str(lay)]= [constElevation for x in range(modDis["cellsperlay"])]
else:
pass
# ### List of arrays of cells and vertex coord
# In[7]:
modDis['vertexEasting'] = np.array([modDis['vertexXmin']+np.sum(modDis['DELR'][:col]) for col in range(modDis['vertexCols'])])
modDis['vertexNorthing'] = np.array([modDis['vertexYmax']-np.sum(modDis['DELC'][:row]) for row in range(modDis['vertexRows'])])
modDis['cellEasting'] = np.array([modDis['vertexXmin']+np.sum(modDis['DELR'][:col])+modDis['DELR'][col]/2 for col in range(modDis['cellCols'])])
modDis['cellNorthing'] = np.array([modDis['vertexYmax']-np.sum(modDis['DELC'][:row])-modDis['DELC'][row]/2 for row in range(modDis['cellRows'])])
# ### Interpolation from Z cell centroid to z vertex
# # Get the BAS Info
# ### Get the grid breakers
# In[8]:
#empty dict to store BAS breakers
modBas['basBreakers']={}
breakerValues = ["INTERNAL","CONSTANT"]
#store the breakers in the dict
lay=0
for item in breakerValues:
for line in basLines:
if item in line:
if 'IBOUND' in line:
modBas['basBreakers']['lay'+str(lay)]=basLines.index(line)
lay+=1
else:
pass
# ### Store ibound per lay
# In[9]:
#empty dict to store cell ibound per layer
modBas['cellIboundList']={}
for lay in range(modDis['cellLays']):
#add auxiliar variables to identify breakers
lineaBreaker = modBas['basBreakers']['lay'+str(lay)]
#two cases in breaker line
if 'INTERNAL' in basLines[lineaBreaker]:
lista = Functions.getListFromBreaker(lineaBreaker,modDis,basLines)
modBas['cellIboundList']['lay'+str(lay)]=lista
elif 'CONSTANT' in basLines[lineaBreaker]:
constElevation = float(disLines[lineaBreaker].split()[1]) #todavia no he probado esto
modBas['cellIboundList']['lay'+str(lay)]= [constElevation for x in range(modDis["cellsperlay"])]
else:
pass
# ### Store Cell Centroids as a Numpy array
# In[10]:
#empty list to store cell centroid
cellCentroidList = []
#numpy array of cell centroid
for row in range(modDis['cellRows']):
for col in range(modDis['cellCols']):
cellCentroidList.append([modDis['cellEasting'][col],modDis['cellNorthing'][row]])
#store cell centroids as numpy array
modDis['cellCentroids']=np.asarray(cellCentroidList)
# ### Grid of XYZ Vertex Coordinates
# In[11]:
modDis['vertexXgrid'] = np.repeat(modDis['vertexEasting'].reshape(modDis['vertexCols'],1),modDis['vertexRows'],axis=1).T
modDis['vertexYgrid'] = np.repeat(modDis['vertexNorthing'],modDis['vertexCols']).reshape(modDis['vertexRows'],modDis['vertexCols'])
modDis['vertexZGrid'] = Functions.interpolateCelltoVertex(modDis,'cellCentroidZList')
# # Get the HDS Info
# ### Get the grid breakers
# In[12]:
#empty dict to store HDS breakers
modFhd['fhdBreakers']={}
#store the breakers in the dict
lay=0
for line in fhdLines:
if 'HEAD' in line:
modFhd['fhdBreakers']['lay'+str(lay)]=fhdLines.index(line)
lay+=1
else:
pass
# ### Store heads per lay
# In[13]:
#empty dict to store cell heads per layer
modFhd['cellHeadGrid']={}
for lay in range(modDis['cellLays']):
#add auxiliar variables to identify breakers
lineaBreaker = modFhd['fhdBreakers']['lay'+str(lay)]
lista = Functions.getListFromBreaker(lineaBreaker,modDis,fhdLines)
array = np.asarray(lista).reshape(modDis['cellRows'],modDis['cellCols'])
modFhd['cellHeadGrid']['lay'+str(lay)]=array
# ### Arrange to transform cell centroid heads to vertex heads
# In[14]:
#empty temporal dictionary to store transformed heads
vertexHeadGridCentroid = {}
#arrange to hace positive heads in all vertex of an active cell
for lay in range(modDis['cellLays']):
matrix = np.zeros([modDis['vertexRows'],modDis['vertexCols']])
for row in range(modDis['cellRows']):
for col in range(modDis['cellCols']):
headLay = modFhd['cellHeadGrid']['lay'+str(lay)]
neighcartesianlist = [headLay[row-1,col-1],headLay[row-1,col],headLay[row,col-1],headLay[row,col]]
headList = []
for item in neighcartesianlist:
if item > -1e+20:
headList.append(item)
if len(headList) > 0:
headMean = sum(headList)/len(headList)
else:
headMean = -1e+20
matrix[row,col]=headMean
matrix[-1,:-1] = modFhd['cellHeadGrid']['lay'+str(lay)][-1,:]
matrix[:-1,-1] = modFhd['cellHeadGrid']['lay'+str(lay)][:,-1]
matrix[-1,-1] = modFhd['cellHeadGrid']['lay'+str(lay)][-1,-1]
vertexHeadGridCentroid['lay'+str(lay)]=matrix
# In[15]:
modFhd['vertexHeadGrid'] = {}
for lay in range(modDis['vertexLays']):
anyGrid = vertexHeadGridCentroid
if lay == modDis['cellLays']:
modFhd['vertexHeadGrid']['lay'+str(lay)] = anyGrid['lay'+str(lay-1)]
elif lay == 0:
modFhd['vertexHeadGrid']['lay0'] = anyGrid['lay0']
else:
value = np.where(anyGrid['lay'+str(lay)]>-1e+20,
anyGrid['lay'+str(lay)],
(anyGrid['lay'+str(lay-1)] + anyGrid['lay'+str(lay)])/2
)
modFhd['vertexHeadGrid']['lay'+str(lay)] = value
# # Get the DRN Info
# In[16]:
### Get the list of drain cells with attributes
# In[17]:
modDrn['DrainNumber'] = int(drnLines[2].split()[0])
modDrn['DrainCells'] = []
for item in range(modDrn['DrainNumber']-4):
anyLine = drnLines[item + 4].split()
anyList = [ int(anyLine[0]), int(anyLine[1]), int(anyLine[2]), float(anyLine[3]), float(anyLine[4])]
modDrn['DrainCells'].append(anyList)
# # Water Tables on Cell and Vextex
# ### Water Table on Cell
# In[18]:
#empty numpy array for the water table
waterTableCellGrid = np.zeros((modDis['cellRows'],modDis['cellCols']))
#obtain the first positive or real head from the head array
for row in range(modDis['cellRows']):
for col in range(modDis['cellCols']):
anyList = []
for lay in range(modDis['cellLays']):
anyList.append(modFhd['cellHeadGrid']['lay' + str(lay)][row,col])
a = np.asarray(anyList)
if list(a[a>-1e+10]) != []: #just in case there are some inactive zones
waterTableCellGrid[row,col] = a[a>-1e+10][0]
else: waterTableCellGrid[row,col] = -1e+10
# In[19]:
### Water Table on Vertex
# In[20]:
#empty numpy array for the water table
waterTableVertexGrid = np.zeros((modDis['vertexRows'],modDis['vertexCols']))
#obtain the first positive or real head from the head array
for row in range(modDis['vertexRows']):
for col in range(modDis['vertexCols']):
anyList = []
for lay in range(modDis['cellLays']):
anyList.append(modFhd['vertexHeadGrid']['lay' + str(lay)][row,col])
a = np.asarray(anyList)
if list(a[a>-1e+10]) != []: #just in case there are some inactive zones
waterTableVertexGrid[row,col] = a[a>-1e+10][0]
else: waterTableVertexGrid[row,col] = -1e+10
# ### Water Table on Cells and Vertex as List
# In[21]:
listWaterTableCell = list(waterTableCellGrid.flatten())
waterTableListVertex = list(waterTableVertexGrid.flatten())
# # Lists for the VTK file
# ### Definition of xyz points for all vertex
# In[22]:
#empty list to store all vertex XYZ
vertexXYZPoints = []
#definition of xyz points for all vertex
for lay in range(modDis['vertexLays']):
for row in range(modDis['vertexRows']):
for col in range(modDis['vertexCols']):
xyz=[
modDis['vertexEasting'][col],
modDis['vertexNorthing'][row],
modDis['vertexZGrid']['lay'+str(lay)][row, col]
]
vertexXYZPoints.append(xyz)
# ### Definition of xyz points for Water Table
# In[23]:
#empty list to store all vertex Water Table XYZ
vertexWaterTableXYZPoints = []
#definition of xyz points for all vertex
for row in range(modDis['vertexRows']):
for col in range(modDis['vertexCols']):
if waterTableVertexGrid[row, col] > -1e+10:
waterTable = waterTableVertexGrid[row, col]
else:
waterTable = 1000
xyz=[
modDis['vertexEasting'][col],
modDis['vertexNorthing'][row],
waterTable
]
vertexWaterTableXYZPoints.append(xyz)
# ### Definition of xyz points for Drain Cells
# In[24]:
#empty list to store drain vertex XYZ list
listDrainCellQuadXYZPoints = []
#definition of xyz points for all drain vertex
for row in range(modDis['vertexRows']):
for col in range(modDis['vertexCols']):
xyz=[
modDis['vertexEasting'][col],
modDis['vertexNorthing'][row],
modDis['vertexZGrid']['lay0'][row,col]
]
listDrainCellQuadXYZPoints.append(xyz)
# ### Definition of IBound list for all cell
# In[25]:
#empty list to store all ibound
listIBound = []
#definition of IBOUND
for lay in range(modDis['cellLays']):
for item in modBas['cellIboundList']['lay'+str(lay)]:
listIBound.append(item)
# ### Definition of all Cell Heads and Vertex Heads
# In[26]:
#empty list to store all cell heads
listCellHead = []
#definition of cellHead
for lay in range(modDis['cellLays']):
for item in list(modFhd['cellHeadGrid']['lay'+str(lay)].flatten()):
listCellHead.append(item)
# In[27]:
#empty list to store all vertex heads
listVertexHead = []
#definition of vertexHead
for lay in range(modDis['vertexLays']):
for item in list(modFhd['vertexHeadGrid']['lay'+str(lay)].flatten()):
listVertexHead.append(item)
# ### Definition of Cell Ibound List
# In[28]:
#empty list to store drain cells IO
listDrainsCellsIO = []
#definition of drain cells
modDrn['DrainCellsGrid'] = np.zeros((modDis['cellRows'],modDis['cellCols']))
for item in modDrn['DrainCells']:
modDrn['DrainCellsGrid'][item[1],item[2]] = 1
listDrainsCellsIO = list(modDrn['DrainCellsGrid'].flatten())
# # Hexahedrons and Quads sequences for the VTK File
# ### List of Layer Quad Sequences (Works only for a single layer)
# In[29]:
#empty list to store cell coordinates
listLayerQuadSequence = []
#definition of hexahedrons cell coordinates
for row in range(modDis['cellRows']):
for col in range(modDis['cellCols']):
pt0 = modDis['vertexCols']*(row+1)+col
pt1 = modDis['vertexCols']*(row+1)+col+1
pt2 = modDis['vertexCols']*(row)+col+1
pt3 = modDis['vertexCols']*(row)+col
anyList = [pt0,pt1,pt2,pt3]
listLayerQuadSequence.append(anyList)
# ### List of Hexa Sequences
# In[30]:
#empty list to store cell coordinates
listHexaSequence = []
#definition of hexahedrons cell coordinates
for lay in range(modDis['cellLays']):
for row in range(modDis['cellRows']):
for col in range(modDis['cellCols']):
pt0 = modDis['vertexperlay']*(lay+1)+modDis['vertexCols']*(row+1)+col
pt1 = modDis['vertexperlay']*(lay+1)+modDis['vertexCols']*(row+1)+col+1
pt2 = modDis['vertexperlay']*(lay+1)+modDis['vertexCols']*(row)+col+1
pt3 = modDis['vertexperlay']*(lay+1)+modDis['vertexCols']*(row)+col
pt4 = modDis['vertexperlay']*(lay)+modDis['vertexCols']*(row+1)+col
pt5 = modDis['vertexperlay']*(lay)+modDis['vertexCols']*(row+1)+col+1
pt6 = modDis['vertexperlay']*(lay)+modDis['vertexCols']*(row)+col+1
pt7 = modDis['vertexperlay']*(lay)+modDis['vertexCols']*(row)+col
anyList = [pt0,pt1,pt2,pt3,pt4,pt5,pt6,pt7]
listHexaSequence.append(anyList)
# # Active Cells Hexahedrons and Quads
# ### Cell Heads and Hexa Sequences
# In[31]:
listHexaSequenceDef = []
listCellHeadDef = []
for i in range(len(listCellHead)):
if listCellHead[i] > -1e-10:
listHexaSequenceDef.append(listHexaSequence[i])
listCellHeadDef.append(listCellHead[i])
# ### Active Cells and Hexa Sequences
# In[32]:
listActiveHexaSequenceDef = []
listIBoundDef = []
#filter hexahedrons and heads for active cells
for i in range(len(listIBound)):
if listIBound[i] > 0:
listActiveHexaSequenceDef.append(listHexaSequence[i])
listIBoundDef.append(listIBound[i])
# ### Water Table Quads and Cells
# In[33]:
listWaterTableQuadSequenceDef = []
listWaterTableCellDef = []
for item in range(len(listWaterTableCell)):
if listWaterTableCell[item] > -1e10:
listWaterTableQuadSequenceDef.append(listLayerQuadSequence[item])
listWaterTableCellDef.append(listWaterTableCell[item])
# ### Drain Cells Quads and Cells
# In[34]:
# Get the secuence and values for cell with drains
listDrainsCellsIODef = []
listDrainsCellsSecuenceDef = []
for item in range(len(listDrainsCellsIO)):
if listDrainsCellsIO[item] > 0:
listDrainsCellsIODef.append(listDrainsCellsIO[item])
listDrainsCellsSecuenceDef.append(listLayerQuadSequence[item])
# # VTK creation
# ### Summary of lists for the vtk creation
# In[35]:
### Point sets
# vertexXYZPoints for XYZ in all cell vertex
# vertexWaterTableXYZPoints for XYZ in all water table quad vertex
# listDrainCellQuadXYZPoints for XYZ in all drain cells quad vertex
### Quad and Hexa secuences
# listHexaSequenceDef for Head Hexa Sequence in all active cells
# listActiveHexaSequenceDef for Active Hexa Sequence in all active cells
# listWaterTableQuadSequenceDef for Water Table Quad Sequence in all active cells
# listDrainsCellsSecuenceDef for Drain Cell Quad Sequence in drain cells
### Cell data
# listCellHeadDef for filtered active cells
# listIBoundDef
# listWaterTableCellDef for filtered water table cells
# listDrainsCellsIODef for filtered drains cells
### Point data
# listVertexHead for heads in all cells
# ### Heads on Vertex and Cells VTK
# In[36]:
textoVtk = open('VTUFiles/VTU_Heads.vtu','w')
#add header
textoVtk.write('<VTKFile type="UnstructuredGrid" version="1.0" byte_order="LittleEndian" header_type="UInt64">\n')
textoVtk.write(' <UnstructuredGrid>\n')
textoVtk.write(' <Piece NumberOfPoints="'+str(len(vertexXYZPoints))+'" NumberOfCells="'+str(len(listHexaSequenceDef))+'">\n')
#point data
textoVtk.write(' <PointData Scalars="Heads">\n')
textoVtk.write(' <DataArray type="Float64" Name="Heads" format="ascii">\n')
for item in range(len(listVertexHead)):
textvalue = str(listVertexHead[item])
if item == 0:
textoVtk.write(' ' + textvalue + ' ')
elif item % 20 == 0:
textoVtk.write(textvalue + '\n ')
else:
textoVtk.write(textvalue + ' ')
textoVtk.write('\n')
textoVtk.write(' </DataArray>\n')
textoVtk.write(' </PointData>\n')
#cell data
textoVtk.write(' <CellData Scalars="Model">\n')
textoVtk.write(' <DataArray type="Float64" Name="CellHead" format="ascii" RangeMin="1" RangeMax="1">\n')
for item in range(len(listCellHeadDef)): #cell list
textvalue = str(int(listCellHeadDef[item]))
if item == 0:
textoVtk.write(' ' + textvalue + ' ')
elif item % 20 == 0:
textoVtk.write(textvalue + '\n ')
else:
textoVtk.write(textvalue + ' ')
textoVtk.write('\n')
textoVtk.write(' </DataArray>\n')
textoVtk.write(' </CellData>\n')
#points definition
textoVtk.write(' <Points>\n')
textoVtk.write(' <DataArray type="Float64" Name="Points" NumberOfComponents="3" format="ascii">\n')
for item in range(len(vertexXYZPoints)):
tuplevalue = tuple(vertexXYZPoints[item])
if item == 0:
textoVtk.write(" %.2f %.2f %.2f " % tuplevalue)
elif item % 4 == 0:
textoVtk.write("%.2f %.2f %.2f \n " % tuplevalue)
elif item == len(vertexXYZPoints)-1:
textoVtk.write("%.2f %.2f %.2f \n" % tuplevalue)
else:
textoVtk.write("%.2f %.2f %.2f " % tuplevalue)
textoVtk.write(' </DataArray>\n')
textoVtk.write(' </Points>\n')
#cell connectivity
textoVtk.write(' <Cells>\n')
textoVtk.write(' <DataArray type="Int64" Name="connectivity" format="ascii">\n')
for item in range(len(listHexaSequenceDef)):
textoVtk.write(' ')
textoVtk.write('%s %s %s %s %s %s %s %s \n' % tuple(listHexaSequenceDef[item]))
textoVtk.write(' </DataArray>\n')
#cell offset
textoVtk.write(' <DataArray type="Int64" Name="offsets" format="ascii">\n')
for item in range(len(listHexaSequenceDef)):
offset = str((item+1)*8)
if item == 0:
textoVtk.write(' ' + offset + ' ')
elif item % 20 == 0:
textoVtk.write(offset + ' \n ')
elif item == len(listHexaSequenceDef)-1:
textoVtk.write(offset + ' \n')
else:
textoVtk.write(offset + ' ')
textoVtk.write(' </DataArray>\n')
#cell type
textoVtk.write(' <DataArray type="UInt8" Name="types" format="ascii">\n')
for item in range(len(listHexaSequenceDef)):
if item == 0:
textoVtk.write(' ' + '12 ')
elif item % 20 == 0:
textoVtk.write('12 \n ')
elif item == len(listHexaSequenceDef)-1:
textoVtk.write('12 \n')
else:
textoVtk.write('12 ')
textoVtk.write(' </DataArray>\n')
textoVtk.write(' </Cells>\n')
#footer
textoVtk.write(' </Piece>\n')
textoVtk.write(' </UnstructuredGrid>\n')
textoVtk.write('</VTKFile>\n')
textoVtk.close()
# ### Active Cell VTK
# In[37]:
textoVtk = open('VTUFiles/VTU_Active.vtu','w')
#add header
textoVtk.write('<VTKFile type="UnstructuredGrid" version="1.0" byte_order="LittleEndian" header_type="UInt64">\n')
textoVtk.write(' <UnstructuredGrid>\n')
textoVtk.write(' <Piece NumberOfPoints="'+str(len(vertexXYZPoints))+'" NumberOfCells="'+str(len(listActiveHexaSequenceDef))+'">\n')
#cell data
textoVtk.write(' <CellData Scalars="Model">\n')
textoVtk.write(' <DataArray type="Int32" Name="Active" format="ascii">\n')
for item in range(len(listIBoundDef)): #cell list
textvalue = str(int(listIBoundDef[item]))
if item == 0:
textoVtk.write(' ' + textvalue + ' ')
elif item % 20 == 0:
textoVtk.write(textvalue + '\n ')
else:
textoVtk.write(textvalue + ' ')
textoVtk.write('\n')
textoVtk.write(' </DataArray>\n')
textoVtk.write(' </CellData>\n')
#points definition
textoVtk.write(' <Points>\n')
textoVtk.write(' <DataArray type="Float64" Name="Points" NumberOfComponents="3" format="ascii">\n')
for item in range(len(vertexXYZPoints)):
tuplevalue = tuple(vertexXYZPoints[item])
if item == 0:
textoVtk.write(" %.2f %.2f %.2f " % tuplevalue)
elif item % 4 == 0:
textoVtk.write('%.2f %.2f %.2f \n ' % tuplevalue)
elif item == len(vertexXYZPoints)-1:
textoVtk.write("%.2f %.2f %.2f \n" % tuplevalue)
else:
textoVtk.write("%.2f %.2f %.2f " % tuplevalue)
textoVtk.write(' </DataArray>\n')
textoVtk.write(' </Points>\n')
#cell connectivity
textoVtk.write(' <Cells>\n')
textoVtk.write(' <DataArray type="Int64" Name="connectivity" format="ascii">\n')
for item in range(len(listActiveHexaSequenceDef)):
textoVtk.write(' ')
textoVtk.write('%s %s %s %s %s %s %s %s \n' % tuple(listActiveHexaSequenceDef[item]))
textoVtk.write(' </DataArray>\n')
#cell offsets
textoVtk.write(' <DataArray type="Int64" Name="offsets" format="ascii">\n')
for item in range(len(listActiveHexaSequenceDef)):
offset = str((item+1)*8)
if item == 0:
textoVtk.write(' ' + offset + ' ')
elif item % 20 == 0:
textoVtk.write(offset + ' \n ')
elif item == len(listActiveHexaSequenceDef)-1:
textoVtk.write(offset + ' \n')
else:
textoVtk.write(offset + ' ')
textoVtk.write(' </DataArray>\n')
#cell types
textoVtk.write(' <DataArray type="UInt8" Name="types" format="ascii">\n')
for item in range(len(listActiveHexaSequenceDef)):
if item == 0:
textoVtk.write(' ' + '12 ')
elif item % 20 == 0:
textoVtk.write('12 \n ')
elif item == len(listActiveHexaSequenceDef)-1:
textoVtk.write('12 \n')
else:
textoVtk.write('12 ')
textoVtk.write(' </DataArray>\n')
textoVtk.write(' </Cells>\n')
#footer
textoVtk.write(' </Piece>\n')
textoVtk.write(' </UnstructuredGrid>\n')
textoVtk.write('</VTKFile>\n')
textoVtk.close()
# ### Water Table VTK
# In[38]:
textoVtk = open('VTUFiles/VTU_WaterTable.vtu','w')
#add header
textoVtk.write('<VTKFile type="UnstructuredGrid" version="1.0" byte_order="LittleEndian" header_type="UInt64">\n')
textoVtk.write(' <UnstructuredGrid>\n')
textoVtk.write(' <Piece NumberOfPoints="'+str(len(vertexWaterTableXYZPoints))+'" NumberOfCells="'+
str(len(listWaterTableCellDef))+'">\n')
#cell data
textoVtk.write(' <CellData Scalars="Water Table">\n')
textoVtk.write(' <DataArray type="Float64" Name="Heads" format="ascii">\n')
for item in range(len(listWaterTableCellDef)):
textvalue = str(listWaterTableCellDef[item])
if item == 0:
textoVtk.write(' ' + textvalue + ' ')
elif item % 20 == 0:
textoVtk.write(textvalue + '\n ')
else:
textoVtk.write(textvalue + ' ')
textoVtk.write('\n')
textoVtk.write(' </DataArray>\n')
textoVtk.write(' </CellData>\n')
#points definition
textoVtk.write(' <Points>\n')
textoVtk.write(' <DataArray type="Float64" Name="Points" NumberOfComponents="3" format="ascii">\n')
for item in range(len(vertexWaterTableXYZPoints)):
tuplevalue = tuple(vertexWaterTableXYZPoints[item])
if item == 0:
textoVtk.write(" %.2f %.2f %.2f " % tuplevalue)
elif item % 4 == 0:
textoVtk.write('%.2f %.2f %.2f \n ' % tuplevalue)
elif item == len(vertexWaterTableXYZPoints)-1:
textoVtk.write("%.2f %.2f %.2f \n" % tuplevalue)
else:
textoVtk.write("%.2f %.2f %.2f " % tuplevalue)
textoVtk.write(' </DataArray>\n')
textoVtk.write(' </Points>\n')
#cell connectivity
textoVtk.write(' <Cells>\n')
textoVtk.write(' <DataArray type="Int64" Name="connectivity" format="ascii">\n')
for item in range(len(listWaterTableQuadSequenceDef)):
textoVtk.write(' ')
textoVtk.write('%s %s %s %s \n' % tuple(listWaterTableQuadSequenceDef[item]))
textoVtk.write(' </DataArray>\n')
#cell offsets
textoVtk.write(' <DataArray type="Int64" Name="offsets" format="ascii">\n')
for item in range(len(listWaterTableQuadSequenceDef)):
offset = str((item+1)*4)
if item == 0:
textoVtk.write(' ' + offset + ' ')
elif item % 20 == 0:
textoVtk.write(offset + ' \n ')
elif item == len(listWaterTableQuadSequenceDef)-1:
textoVtk.write(offset + ' \n')
else:
textoVtk.write(offset + ' ')
textoVtk.write(' </DataArray>\n')
#cell types
textoVtk.write(' <DataArray type="UInt8" Name="types" format="ascii">\n')
for item in range(len(listWaterTableQuadSequenceDef)):
if item == 0:
textoVtk.write(' ' + '9 ')
elif item % 20 == 0:
textoVtk.write('9 \n ')
elif item == len(listWaterTableQuadSequenceDef)-1:
textoVtk.write('9 \n')
else:
textoVtk.write('9 ')
textoVtk.write(' </DataArray>\n')
textoVtk.write(' </Cells>\n')
#footer
textoVtk.write(' </Piece>\n')
textoVtk.write(' </UnstructuredGrid>\n')
textoVtk.write('</VTKFile>\n')
textoVtk.close()
# # Drain Cell VTK
# In[39]:
textoVtk = open('VTUFiles/VTU_DrainCells.vtu','w')
#add header
textoVtk.write('<VTKFile type="UnstructuredGrid" version="1.0" byte_order="LittleEndian" header_type="UInt64">\n')
textoVtk.write(' <UnstructuredGrid>\n')
textoVtk.write(' <Piece NumberOfPoints="'+str(len(listDrainCellQuadXYZPoints))
+'" NumberOfCells="'+str(len(listDrainsCellsSecuenceDef))+'">\n')
#cell data
textoVtk.write(' <CellData Scalars="Water Table">\n')
textoVtk.write(' <DataArray type="Float64" Name="Heads" format="ascii">\n')
for item in range(len(listDrainsCellsIODef)):
textvalue = str(listDrainsCellsIODef[item])
if item == 0:
textoVtk.write(' ' + textvalue + ' ')
elif item % 20 == 0:
textoVtk.write(textvalue + '\n ')
else:
textoVtk.write(textvalue + ' ')
textoVtk.write('\n')
textoVtk.write(' </DataArray>\n')
textoVtk.write(' </CellData>\n')
#points definition
textoVtk.write(' <Points>\n')
textoVtk.write(' <DataArray type="Float64" Name="Points" NumberOfComponents="3" format="ascii">\n')
for item in range(len(listDrainCellQuadXYZPoints)):
tuplevalue = tuple(listDrainCellQuadXYZPoints[item])
if item == 0:
textoVtk.write(" %.2f %.2f %.2f " % tuplevalue)
elif item % 4 == 0:
textoVtk.write('%.2f %.2f %.2f \n ' % tuplevalue)
elif item == len(listDrainCellQuadXYZPoints)-1:
textoVtk.write("%.2f %.2f %.2f \n" % tuplevalue)
else:
textoVtk.write("%.2f %.2f %.2f " % tuplevalue)
textoVtk.write(' </DataArray>\n')
textoVtk.write(' </Points>\n')
#cell definition
textoVtk.write(' <Cells>\n')
textoVtk.write(' <DataArray type="Int64" Name="connectivity" format="ascii">\n')
for item in range(len(listDrainsCellsSecuenceDef)):
textoVtk.write(' ')
textoVtk.write('%s %s %s %s \n' % tuple(listDrainsCellsSecuenceDef[item]))
textoVtk.write(' </DataArray>\n')
textoVtk.write(' <DataArray type="Int64" Name="offsets" format="ascii">\n')
for item in range(len(listDrainsCellsSecuenceDef)):
offset = str((item+1)*4)
if item == 0:
textoVtk.write(' ' + offset + ' ')
elif item % 20 == 0:
textoVtk.write(offset + ' \n ')
elif item == len(listDrainsCellsSecuenceDef)-1:
textoVtk.write(offset + ' \n')
else:
textoVtk.write(offset + ' ')
textoVtk.write(' </DataArray>\n')
textoVtk.write(' <DataArray type="UInt8" Name="types" format="ascii">\n')
for item in range(len(listDrainsCellsSecuenceDef)):
if item == 0:
textoVtk.write(' ' + '9 ')
elif item % 20 == 0:
textoVtk.write('9 \n ')
elif item == len(listDrainsCellsSecuenceDef)-1:
textoVtk.write('9 \n')
else:
textoVtk.write('9 ')
textoVtk.write(' </DataArray>\n')
textoVtk.write(' </Cells>\n')
#footer
textoVtk.write(' </Piece>\n')
textoVtk.write(' </UnstructuredGrid>\n')
textoVtk.write('</VTKFile>\n')
textoVtk.close()
This is the script for the auxiliary functions.
import math
import numpy as np
from scipy.interpolate import griddata
class Functions:
def __init__(self, name):
self.name = name
def getListFromDEL(initbreaker,disLines,celldim):
if 'CONSTANT' in disLines[initbreaker]:
constElevation = float(disLines[initbreaker].split()[1])
anyLines = [constElevation for x in range(celldim)]
elif 'INTERNAL' in disLines[initbreaker]:
#empty array and number of lines
anyLines = []
#final breaker
finalbreaker = initbreaker+1+math.ceil(celldim/10)
#append to list all items
for linea in range(initbreaker+1,finalbreaker,1):
listaitem = [float(item) for item in disLines[linea].split()]
for item in listaitem: anyLines.append(item)
else:
anylines = []
return np.asarray(anyLines)
def getListFromBreaker(initbreaker,modDis,fileLines):
#empty array and number of lines
anyLines = []
finalbreaker = initbreaker+1+math.ceil(modDis['cellCols']/10)*modDis['cellRows']
#append to list all items
for linea in range(initbreaker+1,finalbreaker,1):
listaitem = [float(item) for item in fileLines[linea].split()]
for item in listaitem: anyLines.append(item)
return anyLines
#function that return a dictionary of z values on the vertex
def interpolateCelltoVertex(modDis,item):
dictZVertex = {}
for lay in modDis[item].keys():
values = np.asarray(modDis[item][lay])
grid_z = griddata(modDis['cellCentroids'], values,
(modDis['vertexXgrid'], modDis['vertexYgrid']),
method='nearest')
dictZVertex[lay]=grid_z
return dictZVertex